Plant Molecular Biology
Photosynthesis - molecularly networked and optimized
Photosynthesis - molecularly networked and optimized
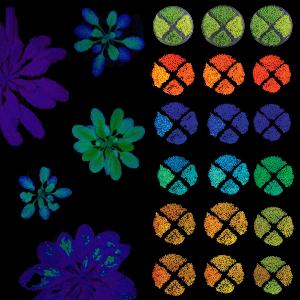
Our primary research interest lies in the in-depth molecular exploration of photosynthesis, and in unraveling its intricate integration with a wide spectrum of cellular processes, both inside and beyond the chloroplast. We closely examine the cellular mechanisms essential for efficient photosynthesis, investigating not only how these functions are controlled within the chloroplast, but also how signalling and metabolic coordination occur between the organelle and the nucleus. Central to our work is a comprehensive, plastid-wide investigation of protein functions, particularly those directly related to photosynthesis, and the regulatory networks orchestrating their activity. To propel advances in photosynthetic engineering, we utilize state-of-the-art synthetic biology and experimental evolution strategies, targeting the redesign and optimization of the photosynthetic machinery. As versatile model systems that are amenable to rapid genetic engineering and precise evolutionary manipulation, we increasingly employ cyanobacterial and algal chassis. These systems provide access to powerful genetic tools and accelerated selection, allowing us to efficiently identify crucial components that contribute to stress adaptation and metabolic efficiency. With this integrated strategy, we strive not only to deepen our fundamental understanding of photosynthetic regulation and cellular integration, but also to pioneer innovative solutions for sustainable agriculture and food security.
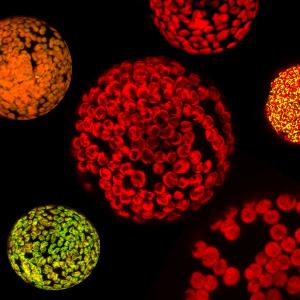
Our multidisciplinary approach brings together genetics, biochemistry, physiology, and molecular biology of plants and other photosynthetic organisms, while also leveraging artificial intelligence, advanced synthetic biology techniques, and laboratory-based evolutionary methodologies in cyanobacteria and algae. By identifying and characterizing key elements underlying adaptation to challenging environmental conditions in these organisms, we can transfer these beneficial traits into higher plants. This endeavor ultimately aims to enhance photosynthetic performance and crop yields under diverse environmental scenarios.
Penzler JF, Naranjo B, Walz S, Marino G, Kleine T, Leister D (2024) A pgr5 suppressor screen un-covers two distinct suppression mechanisms and links cytochrome b6f complex stability to PGR5. Plant Cell 36: 4245-4266. https://doi.org/10.1093/plcell/koae098
Dann M, Ortiz EM, Thomas M, Guljamow A, Lehmann M, Schaefer H, Leister D (2021) Enhancing photosynthesis at high light levels by adaptive laboratory evolution. Nat Plants 7: 681-695. https://doi.org/10.1038/s41477-021-00904-2
Rühle T, Dann M, Reiter B, Schünemann D, Naranjo B, Penzler JF, Kleine T, Leister D (2021) PGRL2 triggers degradation of PGR5 in the absence of PGRL1. Nat Commun 12: 3941. https://doi.org/10.1038/s41477-021-00904-2
Garcia-Molina A, Leister D (2020) Accelerated relaxation of photoprotection impairs biomass accumu-lation in Arabidopsis. Nat Plants 6: 9-12. https://doi.org/10.1038/s41477-019-0572-z
Dann M, Leister D (2019) Evidence that cyanobacterial Sll1217 functions analogously to PGRL1 in enhancing PGR5-dependent cyclic electron flow. Nat Commun 10: 5299. https://doi.org/10.1038/s41467-019-13223-0

